Score-based Generative Modeling of Graphs via the System of Stochastic Differential Equations
Score-based generative models have achieved strong results in image and molecular generation, but applying them to graph-structured data remains challenging due to the complex interplay between node features and graph topology. In this blog, we explore GDSS (Graph Diffusion via the System of Stochastic differential equations), a model that tackles this problem by jointly modeling the evolution of node attributes and adjacency matrices through a system of coupled stochastic differential equations. Using a permutation-equivariant graph neural network and score matching, GDSS generates graphs that maintain both structural validity and semantic coherence.
Introduction: Why Graph Generation Matters
Graphs are fundamental to representing structured relationships in various domains, such as molecular structures in drug discovery
While traditional generative models, such as VAEs, GANs, and autoregressive models, have achieved limited success when applied to graphs due to these complexities, recent advancements in score-based generative modeling
Limitations of Existing Methods
Autoregressive models like GraphRNN
VAE-based models such as GraphVAE
GAN-based models like MolGAN
Flow and early diffusion models like GraphAF
EDP-GNN
While graph generation aims to synthesize graphs that closely match the distribution of observed data, these limitations highlight the need for a model that can jointly model nodes and edges on continuous-time domain, respect graph symmetries, and scale with complexity: and GDSS meets this need.
How GDSS Works
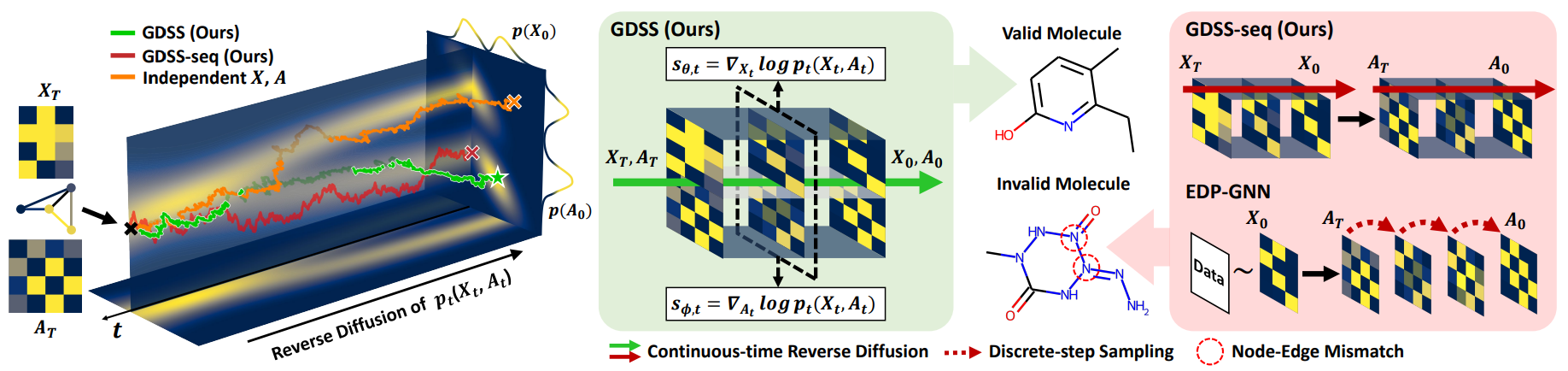
GDSS tackles these limitations by proposing a (1) scored-based graph generation framework (2) on a continuous-time domain that can (3) generate both the node features and the adjacency matrix.
During training, the model learns two score functions that approximate the gradients of the log-density of the noisy data with respect to node features and edges. A permutation-equivariant score based model learns these scores, capturing correlations between structure and semantics. This design allows GDSS to simulate the reverse-time SDEs and sample realistic graphs from noise, achieving state-of-the-art performance on both synthetic and molecular graph benchmarks.
An overview of GDSS’s process is shown below.
0. Data Representation
To start with, GDSS operates on undirected graphs with node features, represented as
$G$ = ($X$, $A$): \(X \in \mathbb{R}^{N \times F}, \quad A \in \mathbb{R}^{N \times N}\),
where $X$ is the node feature matrix for $N$ nodes, each with dimension $F$ features and $A$ is the symmetric adjacency matrix.
1. Forward Diffusion via Coupled SDEs
To model the dependency between $X$ and $A$, GDSS formulates a forward diffusion process of graphs that transforms both of them to a simple noise distribution. The forward process \({G_t = (X_t,A_t)}, t∈[0,T]\) is defined by two independent SDEs, as shown below:
\[d\mathbf{G}_t = \mathbf{f}_t(\mathbf{G}_t) \, dt + \mathbf{g}_t(\mathbf{G}_t) \, d\mathbf{w}, \quad \mathbf{G}_0 \sim p_{\text{data}}\]where $\mathbf{f}_t$ is the linear draft coefficient, $\mathbf{g}_t$ is the diffusion coefficient and $w$ is standard Wiener processes. $\mathbf{f}_t$ and $\mathbf{g}_t$ are chosen such that at the terminal time horizon $T$, the diffused sample $G_T$ approximately follows a prior distribution that has a tractable form to efficiently generate the samples, such as Gaussian distribution. To make it simpler, \(\mathbf{g}_t(\mathbf{G}_t)\) is chosen to be a scalar function $g_t$.
2. Reverse Process with Learned Score Functions
To generate graphs that follow the data distribution, GDSS start from samples of the prior distribution and leverage the following g reverse-time SDE
where $p_t$ is the marginal distribution under the forward diffusion process at time \(t\), \(\bar{\mathbf{w}}\) is a reverse-time standard Wiener process, and \(d\bar{t}\) is an infinitesimal negative time step.
For efficient computing of \(\nabla_{G_t} \log p_t(G_t)\), GDSS utilizes below equations, which operate the same function as above equation. This is a novel approach that interprets the diffusion of a graph as the diffusion of each component that are interrelated through time.
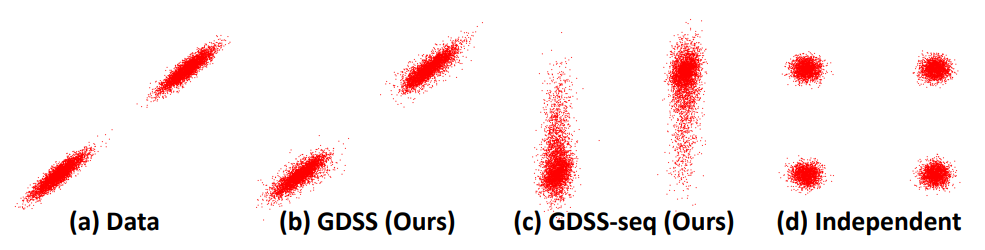
\[dX_t = [f_{1, t}(X_t)-g_{1, t}^2 \nabla_{X_t} \log p_t(X_t, A_t)] d\bar{t} + g_{1, t} d\bar{\mathbf{w}}_1\] \[dA_t = [f_{2, t}(A_t)-g_{2, t}^2 \nabla_{A_t} \log p_t(X_t, A_t)] d\bar{t} + g_{2, t} d\bar{\mathbf{w}}_2\]The key property of GDSS is that the diffusion processes in the system are dependent on each other, as shown in the partial score functions, \(\nabla_{X_t} \log p_t(X_t, A_t)\) and \(\nabla_{A_t} \log p_t(X_t, A_t)\). To show the importance of modeling the dependency, Figure 2 compares the generated samples with the data distribution as bivaraite Gaussian mixture, which shows that GDSS successfully handles correlation of two variables while others ((c) GDSS-seq : that generates \(X\) and \(A\) sequentially and not simultaneously or (d) that ignores the diffusion process of \(X\)) can’t.
Now, since the exact score functions $\nabla_{X_t} \log p_t$ and $\nabla_{A_t} \log p_t$ are unknown, GDSS learns parameterized approximations as follow:
\(s_{\theta, t}(G_t) \approx \nabla_{X_t} \log p_t(G_t), \quad s_A(\phi, t) \approx \nabla_{A_t} \log p_t(G_t)\).
Considering that the score-based models should be trained to minimize the distance to the corresponding ground-truth partial scores, GDSS utilizes a novel objectives that generalize the score matching
However, in above equation, the ground-truth partial scores are not accessible, so GDSS denoises score matching to the partial scores, as shown below:
\[\min_{\theta} \, \mathbb{E}_t \left\{ \lambda_1(t) \, \mathbb{E}_{G_0} \, \mathbb{E}_{G_t|G_0} \left\| s_{\theta,t}(G_t) - \nabla_{X_t} \log p_{0t}(G_t | G_0) \right\|_2^2 \right\}\] \[\min_{\phi} \, \mathbb{E}_t \left\{ \lambda_2(t) \, \mathbb{E}_{G_0} \, \mathbb{E}_{G_t|G_0} \left\| s_{\phi,t}(G_t) - \nabla_{A_t} \log p_{0t}(G_t | G_0) \right\|_2^2 \right\}\]Furthermore, since the drift coefficient of the forward diffusion process is linear as shown below,
\[d\mathbf{G}_t = \mathbf{f}_t(\mathbf{G}_t) \, dt + \mathbf{g}_t(\mathbf{G}_t) \, d\mathbf{w}, \quad \mathbf{G}_0 \sim p_{\text{data}}\]we can get
\[p_{0t}(G_t \mid G_0) = p_{0t}(X_t \mid X_0) \cdot p_{0t}(A_t \mid A_0)\]As a result, the final equation can be summarized as below, with more details for the derivations found in GDSS
Now, it is important to note that estimating the partial scores of GDSS is different from estimating marginal scores \(\nabla_{X_t} \log p_t(X_t) \quad \text{or} \quad \nabla_{A_t} \log p_t(A_t)\) used in previous score-based generative models because GDSS requires capturing the dependency between node features $X_t$ and adjacency matrices $A_t$ through their joint probability over time. Since above training objectives enable effective estimation of these partial scores, the next step is to design model architectures capable of learning them.
3. Permuation-equivariant Score-based Model
For this, GDSS propose two neural architectures based on Graph Neural Networks (GNNs).
For the adjacency score, the model \(s_{\phi,t}(G_t)\) estimates the gradient \(\nabla_{A_t} \log p_t(X_t, A_t).\) This is done by graph multi-head (GMH) attention, which helps identify key relational patterns between nodes, along with higher-order adjacency matrices to account for long-range interactions. These representations are then passed through a multi-layer perceptron (MLP) to produce the final score.
For the node feature score, the model \(s_{\theta,t}(G_t)\) targets \(\nabla_{X_t} \log p_t(X_t, A_t).\) It stacks multiple GNN layers to compute contextual node embeddings from the adjacency matrix, which are again aggregated via an MLP to form the score estimate.
Importantly, both models incorporate time conditioning by scaling their outputs with the standard deviation of the diffusion process at time
\(t,\)
following previous work
4. Solving the Reverse Time SDE System with S4
Once GDSS learns the partial scores for nodes and edges, the next challenge is generating graphs by solving the reverse-time diffusion process:
\[\begin{cases} \mathrm{d}\mathbf{X}_t = f_{1,t}(\mathbf{X}_t)\,\mathrm{d}t + g_{1,t}\,\mathrm{d}\bar{\mathbf{w}}_1 \underbrace{-\, g_{1,t}^2\, s_{\theta,t}(\mathbf{X}_t, \mathbf{A}_t)\,\mathrm{d}t}_{\textbf{S}} \\ \mathrm{d}\mathbf{A}_t = f_{2,t}(\mathbf{A}_t)\,\mathrm{d}t + g_{2,t}\,\mathrm{d}\bar{\mathbf{w}}_2 \underbrace{-\, g_{2,t}^2\, s_{\phi,t}(\mathbf{X}_t, \mathbf{A}_t)\,\mathrm{d}t}_{\textbf{S}} \end{cases} \quad \underbrace{\text{First two terms}}_{\textbf{F}}\]These coupled SDEs are not trivial to solve due to the tight interdependence between $X_t$ and $A_t$. To address this, GDSS introduces S4 (Symmetric Splitting for Systems of SDEs), a new solver that balances accuracy and efficiency by leveraging operator splitting techniques. The method is inspired by symmetric splitting samplers
Each reverse-time step is decomposed into three phases:
- Score Computation: Estimate partial scores \(s_{\theta,t}\) and \(s_{\phi,t}\) using the trained networks.
-
Correction: Use a Langevin MCMC
step to refine the current state \(G_t\), ensuring alignment with the learned score. -
Prediction: Use the Fokker-Planck operators for both drift (F) and score (S) terms in above equation to evolve the graph state backward. This is done via the Trotter splitting
scheme:
where the F-terms correspond to the dynamics of the forward diffusion process and can be sampled exactly from the known transition distribution. However, the S-term which represents score-based updates, is not analytically tractable and is approximated using a simple Euler method. This symmetric arrangement ensures second-order accuracy and reduces numerical artifacts that can accumulate during simulation. Further details regarding the derivations can be found in
Compared to traditional predictor-corrector samplers, S4 cuts the number of score network evaluations by half, achieving a significant reduction in compute without sacrificing fidelity. This design choice is critical because score network evaluation is typically the dominant computational bottleneck.
Moreover, S4 generalizes to systems involving different types of SDEs, including Variance Exploding (VE) and Variance Preserving (VP) forms
To further increase accuracy (at higher computational cost), it is possible to use higher-order integrators like the Runge–Kutta method or substitute Hamiltonian Monte Carlo (HMC)
Experimental Results
Generic Graph Generation
To evaluate the ability of GDSS to generate graphs that match real-world and synthetic graph distributions, a series of experiments on four generic datasets are conducted. These datasets include a mix of small-scale, synthetic, and protein-structured graphs with diverse topologies and statistical properties:
-
Ego-small: 200 ego-centric graphs extracted from the Citeseer citation network
. - Community-small: 100 synthetic graphs constructed from stochastic block models, each representing community structures.
-
Enzymes: 587 protein tertiary structure graphs from the BRENDA database
. - Grid: 100 standard 2D grid graphs, known to be challenging due to their regular, structured nature.
The goal is to assess how well GDSS can learn the distribution of these datasets and generate samples with similar structural properties. Following standard graph generation benchmarks
The similarity between generated and real distributions is quantified using maximum mean discrepancy (MMD), computed with a Gaussian Earth Mover’s Distance (EMD) kernel, which is more stable and well-defined than total variation distance used in some earlier work
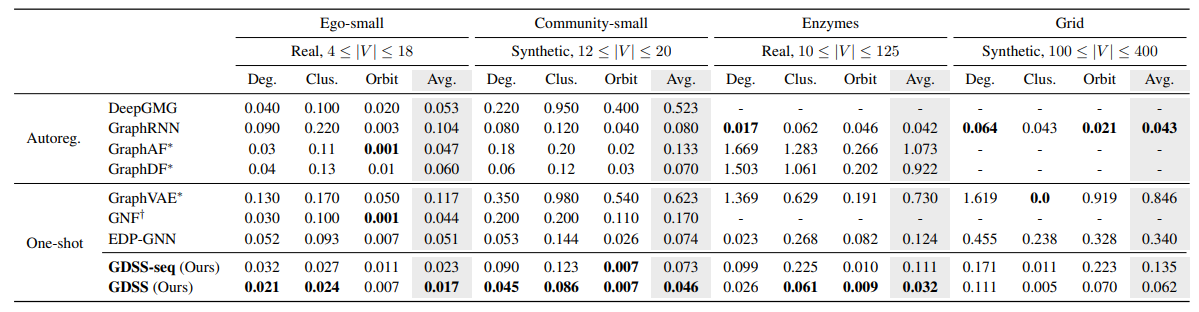
In Table 1, GDSS demonstrates significant improvements over prior one-shot graph generative models, including the previous score-based method EDP-GNN. GDSS not only surpasses all one-shot baselines but also outperforms most autoregressive models, which are typically more expressive but require much higher computational cost.
In particular, on the Grid dataset, GDSS shows comparable the performance to GraphRNN, a strong autoregressive baseline, despite being a one-shot model. By contrast, EDP-GNN completely fails to model such structured graphs, highlighting the limitations of adjacency-only generation and discrete transition dynamics.
Now, one concern in estimating the joint partial score \(\nabla_{A_t} \log p_t(X_t, A_t)\) is that it may appear significantly harder than estimating simpler, marginal-like scores such as \(\nabla_{A_t} \log p_t(X_0, A_t),\) which ignore the time-evolving dependency between node features and edges. Surprisingly, the empirical results indicate the opposite, as shown in Figure 3.
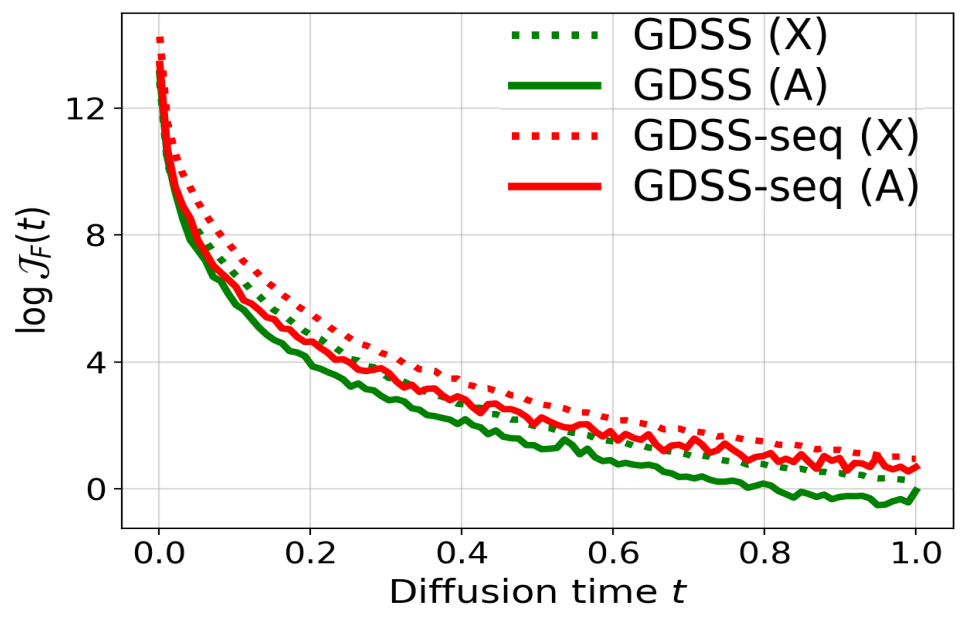
To investigate this, the complexity of score-based models is measured using the squared Frobenius norm of the Jacobians of their score functions, denoted \(J_F(t)\). This metric reflects the sensitivity of the model and the smoothness of the learned score landscape. GDSS is compared against GDSS-seq, a variant that simplifies score estimation by decoupling node and edge interactions over time.
As shown in Figure 3, GDSS consistently exhibits lower model complexity than GDSS-seq when trained on the Ego-small dataset. This holds for both partial score functions:
\[\nabla_{A_t} \log p_t(X_t, A_t) \quad \text{and} \quad \nabla_{X_t} \log p_t(X_t, A_t)\]This counterintuitive result highlights a key strength of GDSS that, learning the true joint dynamics between node features and edges not only improves generative performance (as shown in Table 1), but also simplifies the optimization landscape. The reduced Jacobian norms suggest that joint modeling provides smoother, more learnable gradients, enabling more stable and efficient training.
Molecule Generation Performance and Efficiency
To evaluate molecular graph generation, the experiments are conducted on two widely used datasets:
-
QM9
: A dataset of small organic molecules with up to 9 heavy atoms. -
ZINC250k
: A subset of the ZINC database containing drug-like molecules.
Following prior works
The evaluation focuses on three aspects:
-
Fréchet ChemNet Distance (FCD)
, which computes the distance between generated and real molecules in chemical feature space. -
NSPDK MMD
, a graph-structured MMD metric that captures both node and edge features. -
Validity w/o correction, defined as the fraction of chemically valid molecules generated without any post-processing or valency correction. Notably, this differs from earlier works by allowing atoms with formal charges, following
, to better reflect the diversity of real-world molecular graphs.
All models generate 10,000 molecules, and generation time is measured in terms of RDKit-formatted outputs. Additional implementation details including hyperparameters are described in Appendix C.3 of the paper.
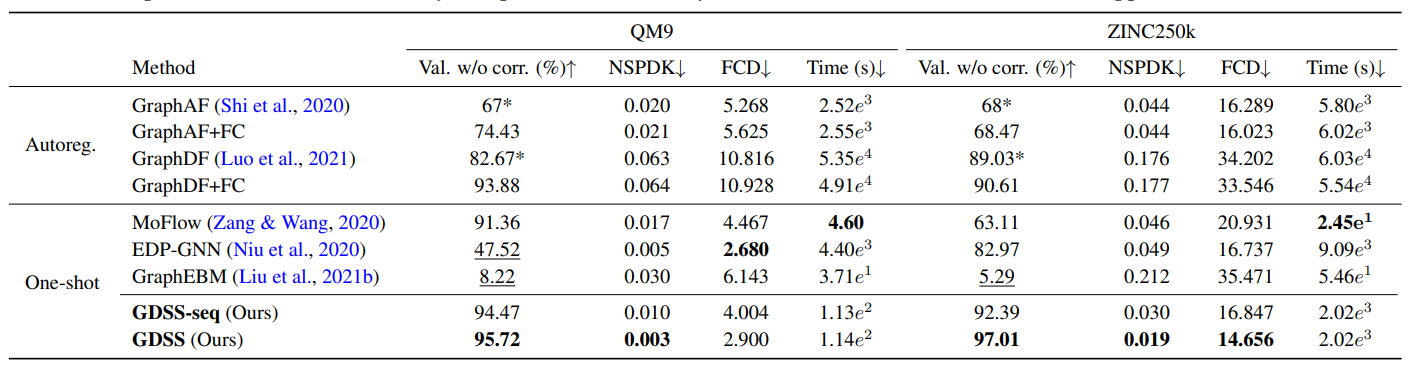
As shown in Table 2, GDSS achieves the highest validity among all methods without relying on post-hoc valency correction. This indicates that GDSS effectively learns the chemical valency rules, which fundamentally rely on modeling the relationship between atoms (nodes) and bonds (edges).
GDSS also significantly outperforms all baselines in NSPDK MMD, and most in Fréchet ChemNet Distance (FCD), demonstrating that the generated molecules are not only structurally realistic in graph space but also chemically plausible in feature space. This shows GDSS’s ability to model complex joint distributions of molecules with diverse node and edge types.
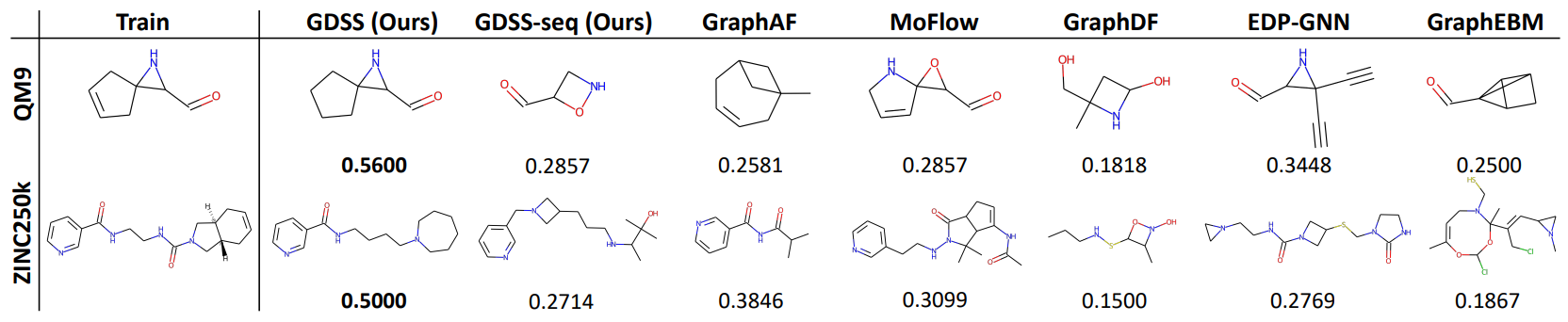
Visualizations in Figure 4 further emphasize this result: molecules generated by GDSS often share large substructures with those from the training set. In contrast, baseline methods frequently produce molecules with less resemblance or completely unrelated structures, highlighting GDSS’s superior generative fidelity.
Beyond quality, GDSS is also highly efficient, as shown in Table 2. On the QM9 dataset, GDSS achieves a 450× speed-up in generation time compared to GraphDF, one of the strongest autoregressive baselines. This efficiency stems from GDSS’s continuous-time diffusion framework, which replaces costly sequential sampling with a more scalable reverse-time SDE simulation.
Additionally, both GDSS and its variant GDSS-seq outperform EDP-GNN in generation speed. This further validates that modeling graph transformation as a continuous diffusion process is not only more expressive but also much more practical than discrete-step perturbation methods.
Ablation Studies
Why Modeling Node-Edge Dependency Matters
To assess the importance of capturing the dependency between node features and edges, GDSS is compared with GDSS-seq, where GDSS models the joint dependency between $X$ and $A$, while GDSS-seq assumes that $A$ depends only on $X$, simplifying the reverse SDE system. This subtle design choice turns out to be crucial.
Across all benchmarks in Table 1 (generic graphs) and Table 2 (molecules), GDSS consistently outperforms GDSS-seq in every metric. The results clearly demonstrate that accurately modeling the graph distribution requires learning both node and edge interactions through time, rather than treating them as partially independent.
For molecule generation in particular, the benefit is measurable in terms of chemical validity. GDSS achieves higher validity scores without needing valency correction, directly showing that it better captures the structure–function relationship between atoms and bonds.
These findings confirm that the proposed system of SDEs—where the partial scores
\[\nabla_{X_t} \log p_t(X_t, A_t)\]and
\[\nabla_{A_t} \log p_t(X_t, A_t)\]are co-trained—provides a more faithful and powerful approach to generative graph modeling.
Significance of S4 Solver
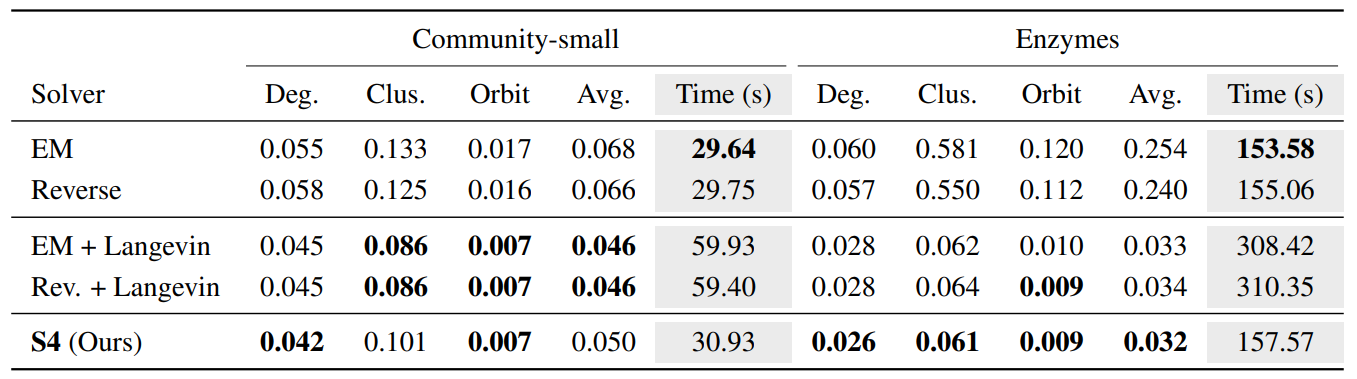
As shown in Table 3, S4 significantly outperforms both Euler–Maruyama (EM) and Reverse sampler, which are simple baseline solvers for reverse-time diffusion, validating the advantage of including both prediction and correction steps. More impressively, despite using half the number of score network evaluations, S4 also outperforms the PC samplers, a more advanced methods using Langevin MCMC for correction.
This efficiency stems from S4’s use of symmetric splitting and score reuse, which allow it to maintain high sampling accuracy while dramatically reducing computational overhead. Since score-based model evaluations dominate the cost of solving stochastic differential equations (SDEs), this reduction is critical for scaling to large or complex graphs. By combining accuracy, efficiency, and generality, S4 becomes a powerful and practical solver for simulating the reverse diffusion process in GDSS.
Personal Thoughts
GDSS redefines the paradigm of graph generation by enabling the joint, continuous, and permutation-invariant synthesis of both graph structure and semantic node features. It uses the stochastic differential equations (SDEs) and the score-based diffusion modeling to navigate the complex space of structured data. By coupling feature-edge diffusion dynamics and employing equivariant GNN-based score estimators, GDSS establishes a framework for generating valid, diverse, and semantically coherent graphs, contributing to areas including, molecular modeling, synthetic biology, and network science.
Key Takeaways
-
Unified Joint Modeling
By treating the adjacency matrix and node features as co-evolving under a system of SDEs, this work overcomes the typical fragmentation in graph generation. It significantly improves structural-semantic alignment, which is crucial for tasks like molecular graph synthesis. -
Permutation-Invariant Score Learning
Leveraging permutation-equivariant score-based model ensures that the model respects the graph’s inherent symmetries, meaning that the node permutations do not affect the outcome, addressing a key limitation in many autoregressive approaches. -
Continuous and Flexible Framework
Diffusion-based generative modeling offers continuous-time flexibility and avoids common drawbacks like mode collapse seen in GANs and VAEs. It is especially suited for high-dimensional, sparse, and structured graph data. -
Empirical Strength
The GDSS model achieves state-of-the-art results on graph generation tasks, outperforming both one-shot and autoregressive baselines on benchmarks like QM9 and ZINC250k in terms of Validity (no correction), NSPDK MMD (structural fidelity), FCD (chemical distribution closeness). Among the SDE types, VE-SDE paired with the efficient S4 solver delivers the best stability, quality, and sampling speed, outperforming existing solvers like EM and PC.
Future Research Directions
Based on the proposed method, I believe that below approaches could be a possible further research direction.
-
Conditional Graph Generation
Extending the framework to support property or topology conditioned generation would broaden its utility in drug discovery, materials design, and graph-based reasoning. -
Learning with Hard Constraints
Incorporating domain-specific constraints, like chemical valency rules, as projection steps or within constraint-aware score functions could significantly improve the validity of generated graphs. -
Dynamic and Temporal Graphs
Extending to dynamic graph generation, where both node features and edges evolve over time, would benefit applications such as interaction networks, financial systems, and physical simulations -
Latent Diffusion for Graphs
Considering latent diffusion models such as Stable Diffusion, I think that exploring generating graphs in a learned latent space could be another exiting research approach to improve efficiency without sacrificing fidelity. -
Multimodal Graph Generation
Graphs often co-occur with other modalities in scientific or social domain, in forms such as text and images. Designing joint diffusion frameworks for cross-modal graph generation could be another possible research direction.
Extending GDSS: Latent Diffusion for Graphs
In this section, I outline a potential extension of GDSS, inspired by recent advances in latent diffusion models such as Stable Diffusion
One promising direction for scaling GDSS is to adapt its reverse-time SDE framework to operate in a latent space, rather than directly on full-resolution graphs. This could be a valid approach since the latent formulation has been effective in other domains for reducing computational cost while preserving high generative fidelity.
We can design the latent diffusion framework for graphs with the following components:
-
Graph Encoder
A permutation-invariant encoder, such as a GNN with hierarchical pooling, maps each graph \(G = (X, A)\) to a compact latent vector \(z \in \mathbb{R}^d,\) capturing both topological and semantic information. -
Latent Diffusion Process
A continuous-time SDE is applied in the latent space. The model learns a score function \(s_{\psi,t}(z)\) to reverse the noising process and generate new latent vectors by simulating reverse-time dynamics. -
Graph Decoder
A decoder reconstructs graph structures from latent representations, possibly using a graph-based VAE, graph transformer, or autoregressive generator conditioned on \(z.\)
This latent-GDSS architecture offers several advantages:
- Efficiency: Operating in the latent space reduces the dimensionality of the generative modeling problem, lowering training and sampling cost.
- Regularization: The encoder-decoder structure encourages a more structured, semantically meaningful manifold, improving generalization.
- Modality Compatibility: A shared latent space can enable multimodal extensions, such as text-to-graph generation or property-conditioned synthesis.
However, a core challenge is ensuring that the latent space remains sufficiently smooth and expressive to support stable diffusion modeling. This could be addressed through contrastive pretraining, joint training with score-based regularization, or denoising objectives that align with the graph decoder.